Introduction
Antibody discovery has revolutionized the pharmaceutical landscape, with monoclonal antibodies now representing the fastest-growing class of therapeutics for treating cancer, autoimmune diseases, and infectious diseases. The journey from identifying a therapeutic target to obtaining a fully characterized, sequenced antibody candidate requires a sophisticated, multi-stage workflow that combines immunology, molecular biology, and cutting-edge analytical technologies.
Antibodies are critical components of the immune system and have become essential tools in research, diagnostics, and therapeutics. The journey from antibody discovery to sequencing involves a series of meticulously coordinated steps that ensure the production of high-quality antibodies tailored for specific applications. This blog outlines the complete workflow, emphasizing the methodologies, technologies, and best practices involved in each phase.
At ResolveMass Laboratories Inc., we understand that successful antibody discovery demands more than just technical capability—it requires strategic planning, rigorous validation, and seamless integration of multiple specialized techniques. This article provides a detailed roadmap through each critical stage of the antibody discovery and sequencing workflow, offering insights gained from years of supporting researchers and biotech companies in their therapeutic development programs.
Summary
Antibody discovery represents the critical first step in developing therapeutic antibodies, diagnostic tools, and research reagents that are transforming modern medicine. This comprehensive guide walks you through the complete workflow from initial target selection to final sequencing, providing actionable insights for researchers and biotech professionals.
Key Takeaways:
- Antibody discovery begins with strategic target identification and validation to ensure therapeutic relevance
- Multiple discovery platforms exist including hybridoma technology, phage display, and single B-cell sequencing
- Immunization strategies and library construction are foundational to generating diverse antibody candidates
- High-throughput screening and functional characterization identify lead candidates with optimal binding properties
- Antibody sequencing provides the genetic blueprint necessary for recombinant production and therapeutic development
- Integration of computational tools and AI is accelerating discovery timelines and improving success rates
- Partnering with experienced laboratories ensures quality, reproducibility, and regulatory compliance
1: Understanding Antibody Discovery: Why It Matters
Antibody discovery is the systematic process of identifying and isolating antibodies that bind specifically to a target of interest. These targets typically include disease-related proteins, cell surface receptors, or pathogenic antigens. The antibodies identified through this process serve as:
- Therapeutic candidates for drug development
- Diagnostic reagents for detecting diseases
- Research tools for understanding biological mechanisms
- Targeting moieties for drug delivery systems
The global therapeutic antibody market exceeded $150 billion in 2023, demonstrating the immense clinical and commercial value of successful antibody discovery programs. However, the path from target to therapeutic is complex, with success rates heavily dependent on the quality and efficiency of the initial discovery workflow.
Stage 1: Target Selection and Validation in Antibody Discovery
The first critical decision in any antibody discovery program is target selection. The target must be disease-relevant, accessible to antibodies, and capable of modulating the desired biological pathway when bound by an antibody.
Key Considerations for Target Selection:
- Disease relevance: Is the target causally linked to disease pathology?
- Druggability: Can antibodies physically access the target in vivo?
- Safety profile: Will targeting this protein cause unacceptable toxicity?
- Competitive landscape: Are existing antibodies already targeting this protein?
1. Antibody Discovery
The first step in the workflow is antibody discovery, where the goal is to identify specific antibodies that bind to target antigens. Various approaches are utilized in this phase, including:
A. Hybridoma Technology
Developed by Georges Köhler and César Milstein in the 1970s, hybridoma technology involves fusing myeloma cells with B lymphocytes from immunized animals (usually mice). This method allows for the production of monoclonal antibodies (mAbs) with high specificity.
- Immunization: Mice or other suitable animals are immunized with the target antigen, leading to the production of B cells that secrete antibodies against the antigen.
- Fusion: The B cells are then fused with myeloma cells to create hybridoma cells that can proliferate indefinitely and produce antibodies.
- Selection: Hybridomas producing antibodies specific to the target antigen are selected through screening assays, such as enzyme-linked immunosorbent assays (ELISA).
B. Phage Display Technology
Phage display is a powerful method for discovering antibodies, particularly in cases where traditional hybridoma technology may not be feasible.
- Library Construction: A diverse library of antibodies is constructed by inserting antibody gene fragments into a bacteriophage’s genome.
- Binding Selection: Phages displaying antibodies that bind to the target antigen are isolated through several rounds of selection, washing, and amplification.
- Characterization: The selected phages are characterized to identify the antibodies they express.
C. Next-Generation Sequencing (NGS)
Advancements in sequencing technology have enabled researchers to utilize NGS for antibody discovery, allowing for a more comprehensive and efficient identification of potential antibodies.
- Single-Cell Sequencing: Isolating single B cells followed by sequencing their antibody genes provides a wealth of information about antibody diversity and specificity.
- High-Throughput Screening: NGS can be combined with high-throughput screening methods to identify potential therapeutic candidates rapidly.
Target Validation Methods:
| Method | Purpose | Timeline |
|---|---|---|
| Bioinformatics analysis | Assess target expression patterns | 1-2 weeks |
| Cell-based assays | Confirm functional relevance | 2-4 weeks |
| Animal models | Validate in vivo significance | 2-6 months |
| Structural studies | Identify epitope accessibility | 1-3 months |
Thorough target validation at this stage prevents costly failures downstream. At ResolveMass Laboratories Inc., we collaborate with clients to assess target feasibility using comprehensive bioinformatics and preliminary functional studies before committing to full discovery campaigns.
Stage 2: Choosing Your Antibody Discovery Platform
Once the target is validated, selecting the appropriate discovery platform is crucial. Each platform offers distinct advantages depending on your specific requirements for antibody format, species origin, and timeline.
Hybridoma Technology for Antibody Discovery
Hybridoma technology remains a proven approach for generating monoclonal antibodies. This method involves immunizing animals (typically mice or rabbits) with the target antigen, isolating B-cells from immunized animals, and fusing these cells with immortal myeloma cells to create hybridomas that continuously produce antibodies.
Advantages:
- Produces high-affinity antibodies through natural immune selection
- Generates fully functional antibodies with proper folding
- Established regulatory pathway for therapeutic development
- No specialized equipment beyond standard cell culture facilities
Considerations:
- Longer timelines (3-6 months)
- Limited to immunogenic targets
- Animal use required
- Restricted to certain species
Phage Display and In Vitro Selection
Phage display revolutionized antibody discovery by enabling selection of antibodies entirely in vitro from large libraries of antibody fragments displayed on bacteriophage surfaces.
Advantages:
- No animal immunization required
- Works with toxic or non-immunogenic targets
- Rapid selection cycles (4-8 weeks)
- Easy genetic manipulation and optimization
Considerations:
- May produce lower initial affinity than immune-derived antibodies
- Requires specialized library construction
- Additional optimization often needed
Single B-Cell Technology
Modern single B-cell sequencing approaches allow direct isolation of antibody sequences from individual B-cells, preserving natural heavy-light chain pairing.
Advantages:
- Rapid discovery (2-4 weeks)
- Captures rare antibodies
- Maintains native pairing
- Enables human antibody discovery
Considerations:
- Requires sophisticated equipment
- Higher upfront costs
- Limited throughput in some formats
Stage 3: Immunization and Library Construction
For immune-based antibody discovery approaches, the immunization strategy significantly impacts the quality of antibody candidates generated.
Optimized Immunization Protocols
Successful immunization requires careful attention to:
- Antigen format: Recombinant protein, peptide, DNA, or whole cells
- Adjuvant selection: Balancing immune stimulation with animal welfare
- Dosing schedule: Optimizing boost timing for affinity maturation
- Route of administration: Subcutaneous, intraperitoneal, or footpad injection
Typical immunization schedules span 6-12 weeks, with multiple boosts to drive affinity maturation through the natural immune response. Monitoring antibody titers throughout immunization helps determine optimal harvest timing.
Library Construction Best Practices
For display technologies, library quality directly determines discovery success. Key parameters include:
- Library diversity: Minimum 10^9-10^11 unique clones for naïve libraries
- Quality control: Sequencing to verify insert integrity and diversity
- Cloning efficiency: Minimizing bias during amplification
- Functional display: Ensuring proper antibody folding on display scaffold
Stage 4: High-Throughput Screening and Selection
Once antibodies are generated (through hybridoma supernatants, phage selections, or B-cell isolation), identifying lead candidates requires systematic screening.
Primary Screening: Binding Identification
Initial screening identifies antibodies that bind to the target antigen. Common methods include:
- ELISA (Enzyme-Linked Immunosorbent Assay): Cost-effective, scalable to hundreds of samples
- Flow cytometry: Confirms binding to cell-surface targets
- Surface plasmon resonance (SPR): Real-time binding kinetics
- Biolayer interferometry (BLI): Label-free affinity measurements
Screening Strategy Tips:
- Use multiple antigen formats to eliminate format-specific binders
- Include negative controls to identify non-specific binders
- Prioritize diverse candidates rather than many similar clones
- Establish clear cut-off criteria before screening begins
Secondary Screening: Functional Characterization
Binding alone is insufficient—antibodies must demonstrate functional activity relevant to the therapeutic or research application.
Functional Assays Include:
- Neutralization assays: Does the antibody block target activity?
- Cell-based assays: Does binding translate to cellular effects?
- Competition assays: Does it block ligand-receptor interactions?
- Internalization studies: For antibody-drug conjugate applications
At this stage, 5-20 lead candidates typically advance to detailed characterization, representing the best balance of binding affinity, specificity, and functional activity.
Stage 5: Lead Optimization and Characterization
Selected lead candidates undergo comprehensive characterization to assess their suitability for the intended application and identify any potential liabilities.
Biophysical Characterization
Understanding antibody biophysical properties is critical for developability:
| Property | Measurement Method | Ideal Range |
|---|---|---|
| Affinity (KD) | SPR, BLI | pM to nM |
| Specificity | Cross-reactivity panel | Target-specific |
| Stability | Thermal shift (Tm) | >65°C |
| Aggregation | SEC, DLS | <5% aggregates |
| Expression yield | Transient expression | >100 mg/L |
Epitope Mapping
Determining where antibodies bind on the target provides insights into:
- Mechanism of action
- Potential for antibody combinations
- Cross-species reactivity
- Competition with existing therapeutics
Epitope mapping approaches:
- Peptide scanning arrays
- Hydrogen-deuterium exchange mass spectrometry (HDX-MS)
- Cryo-electron microscopy (cryo-EM)
- Alanine scanning mutagenesis
Stage 6: Antibody Sequencing—The Gateway to Development
Antibody sequencing represents the critical transition point from discovery to development. Obtaining accurate antibody sequences is essential because:
- Enables recombinant production: Sequences allow antibody production in any expression system
- Supports patent applications: Sequence data provides intellectual property protection
- Facilitates engineering: Sequence information enables affinity maturation and format conversion
- Ensures reproducibility: Eliminates dependence on cell lines that may drift or be lost
Antibody Sequencing Approaches
Mass Spectrometry-Based Sequencing:
Modern mass spectrometry provides comprehensive antibody characterization without requiring live cells or genetic material. At ResolveMass Laboratories Inc., we specialize in advanced mass spectrometry techniques that deliver:
- Complete heavy and light chain sequences
- Identification of post-translational modifications
- Detection of sequence variants within populations
- Verification of complementarity-determining regions (CDRs)
Advantages of MS-based sequencing:
- Works with purified antibodies from any source
- Identifies modifications affecting function
- Provides confident sequence assignment
- No PCR amplification bias
DNA-Based Sequencing:
Traditional Sanger sequencing or next-generation sequencing (NGS) of antibody-encoding genes offers:
- Direct genetic sequence information
- Easy subsequent cloning for recombinant expression
- Cost-effective for large panels
Hybrid Approaches:
Combining DNA sequencing with mass spectrometry confirmation provides the highest confidence, ensuring genetic sequences match the actual antibody protein.
Quality Control in Antibody Sequencing
Rigorous quality control is non-negotiable:
- Multiple overlapping peptides to ensure full coverage
- De novo sequencing to verify against databases
- Functional validation of synthesized antibodies based on sequences
- Comparison with original clone to confirm sequence accuracy
Stage 7: Recombinant Expression and Validation
With sequences in hand, antibodies are cloned into expression vectors and produced recombinantly to confirm that sequenced antibodies retain desired properties.
Expression System Selection
| System | Advantages | Best For |
|---|---|---|
| Mammalian (CHO, HEK293) | Native glycosylation, proper folding | Therapeutic development |
| Bacterial (E. coli) | Rapid, inexpensive | Fragments, research reagents |
| Yeast | Moderate glycosylation, scalable | Diagnostics, some therapeutics |
| Insect cells | Complex proteins | Difficult-to-express antibodies |
Validation Studies
Recombinantly expressed antibodies must demonstrate equivalence to the original discovery clone:
- Binding affinity: Should match original within 2-3 fold
- Functional activity: Must retain neutralization or other functional properties
- Specificity profile: Cross-reactivity panel should be identical
- Biophysical properties: Similar stability and expression levels
This validation step is critical—even single amino acid sequencing errors can dramatically affect antibody properties.
2: Integration of Computational Tools and AI in Antibody Discovery
Modern antibody discovery increasingly leverages computational approaches to accelerate timelines and improve success rates.
Applications of AI and Machine Learning:
- In silico antibody design: Predicting optimal CDR sequences
- Developability prediction: Identifying liability-prone sequences early
- Epitope prediction: Computational modeling of binding sites
- Affinity maturation: Guiding mutations to improve binding
Benefits of computational integration:
- Reduced experimental screening burden
- Earlier identification of developability issues
- Enhanced understanding of structure-function relationships
- Accelerated optimization cycles
At ResolveMass Laboratories Inc., we integrate computational predictions with experimental validation to provide clients with data-driven insights throughout the discovery process.
3: Common Challenges and Solutions in Antibody Discovery Workflows
Challenge 1: Low-Affinity Antibodies
Solutions:
- Extend immunization schedules for greater affinity maturation
- Implement secondary screening focused on affinity ranking
- Apply in vitro affinity maturation techniques
- Consider alternative discovery platforms
Challenge 2: Poor Developability
Solutions:
- Include developability screening earlier in workflow
- Use computational tools to flag problematic sequences
- Optimize formulation conditions
- Consider antibody engineering to improve properties
Challenge 3: Loss of Activity After Sequencing
Solutions:
- Verify sequence accuracy through multiple methods
- Check for critical post-translational modifications
- Optimize expression conditions to match original
- Consider producing in alternative expression systems
Challenge 4: Lack of Functional Activity Despite Binding
Solutions:
- Map epitope to understand binding location
- Test multiple functional assay formats
- Consider generating antibodies to different epitopes
- Evaluate alternative antibody formats (e.g., bispecifics)
4: Regulatory Considerations for Therapeutic Antibody Discovery
For antibodies intended for therapeutic use, regulatory requirements shape discovery workflows from the beginning.
Early-Stage Regulatory Considerations:
- Documentation: Maintain detailed records of all discovery steps
- Animal welfare: Follow IACUC-approved protocols for immunization
- GMP readiness: Plan for eventual manufacturing under GMP conditions
- Intellectual property: Document inventorship and maintain confidentiality
- Sequence validation: Ensure sequences meet regulatory standards for accuracy
IND-Enabling Studies:
Selected therapeutic candidates will eventually require:
- Developability assessments
- Pharmacology studies
- Toxicology studies
- CMC (Chemistry, Manufacturing, Controls) development
Partnering with experienced laboratories that understand these downstream requirements helps avoid discovery decisions that create regulatory challenges later.
Conclusion
Conclusion: Your Partner in Antibody Discovery Success
The journey from antibody discovery to sequencing represents a complex but systematic process that, when executed properly, yields therapeutic candidates and research tools with enormous potential to advance human health. Success requires not just technical capability but strategic planning, quality systems, and deep expertise across multiple disciplines.
The future of antibody therapeutics is bright, with new targets, novel formats, and advanced technologies expanding what’s possible. By understanding and optimizing the complete workflow from antibody discovery through sequencing, you position your program for success in this dynamic and impactful field.
At ResolveMass Laboratories Inc., we are committed to supporting this workflow through our state-of-the-art facilities and expertise in antibody discovery, production, and sequencing.
Frequently Asked Questions:
Antibody Discovery is the process of identifying antibodies that specifically bind to a biological target with high affinity and functional activity.
It forms the foundation of therapeutic antibody development, diagnostics, and research applications.
Antibody sequencing converts a functional antibody into a reproducible and scalable molecular asset.
It enables recombinant expression, intellectual property protection, batch consistency, and regulatory-ready development.
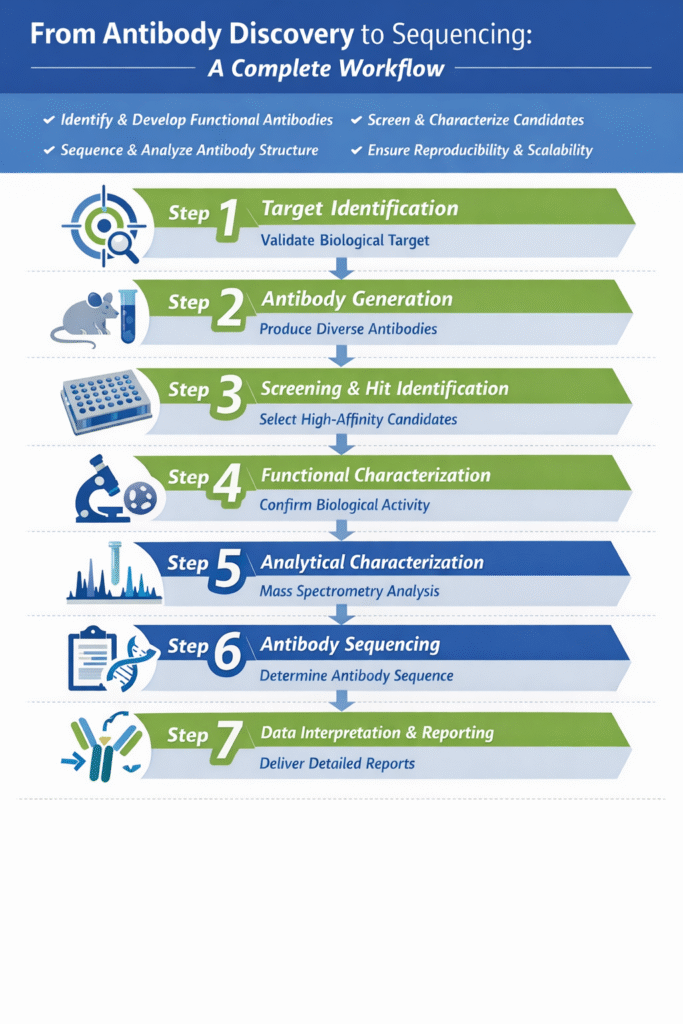
The antibody discovery workflow includes target identification, antibody generation, screening, functional characterization, analytical characterization, sequencing, and data reporting.
Each step builds confidence before advancing to development.
Mass spectrometry enables accurate molecular characterization and sequence determination of antibodies.
Techniques like LC-MS/MS support intact mass analysis, peptide mapping, and de novo antibody sequencing.
Functional characterization confirms that antibody binding leads to the desired biological effect.
This may include neutralization, receptor modulation, or effector function assays.
Modern MS-based antibody sequencing provides high sequence coverage and confidence when combined with expert interpretation.
Orthogonal validation and analytical controls further enhance reliability.
Reference
- Cristina Parola, Daniel Neumeier, Sai T. Reddy.Integrating high-throughput screening and sequencing for monoclonal antibody discovery and engineering.https://onlinelibrary.wiley.com/doi/full/10.1111/imm.12838
- Automated Antibody De Novo Sequencing and Its Utility in Biopharmaceutical Discovery.https://pubs.acs.org/doi/abs/10.1007/s13361-016-1580-0
- Editorial: Next-Generation Sequencing of Human Antibody Repertoires for Exploring B-cell Landscape, Antibody Discovery and Vaccine Development.https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2020.01344/full
- William H. Robinson.Sequencing the functional antibody repertoire—diagnostic and therapeutic discovery.https://www.nature.com/articles/nrrheum.2014.220
- Advances in antibody discovery from human BCR repertoires.https://www.frontiersin.org/journals/bioinformatics/articles/10.3389/fbinf.2022.1044975/full
- Ulla Ravn , Gérard Didelot , Sophie Venet , Kwok-Ting Ng , Franck Gueneau , François Rousseau , Sébastien Calloud , Marie Kosco-Vilbois, Nicolas Fischer.Deep sequencing of phage display libraries to support antibody discovery.https://www.sciencedirect.com/science/article/abs/pii/S1046202313000571